
|
|
Welcome to DynOmics Portal for computing and visualizing biomolecular systems dynamics!
Below is a roadmap for using the different components of our portal. ENM 1.0 provides a unifying user-friendly interface for efficiently performing a broad range of computations by biologists. |
|
Server for all GNM + ANM computations, for evaluating the changes in dynamics in the presence of environment, and for identifying potential functional sites, e.g. key mechanical residues, residues acting as sensors and effectors derived from perturbation response analysis, residues, signaling sites potentially involved in allosteric communication, construction of all atom conformers along ANM modes, animations
Database of GNM dynamics pre-computed for all PDB structures (data on mean-square fluctuations, B-factors, domain separations, correlations between domain movements for biological assemblies)
API for analyzing the structure-based dynamics of biomolecular systems, including the ensemble and/or family properties of sets of conformers or structural homologs, and conformational sampling using elastic network models (ANM and GNM), bridging with sequence conservation and evolution and target druggability
Server for visualizing and downloading animations of collective modes, movies, inter-node distance fluctuations, correlations, deformation energy, based on anisotropic network model
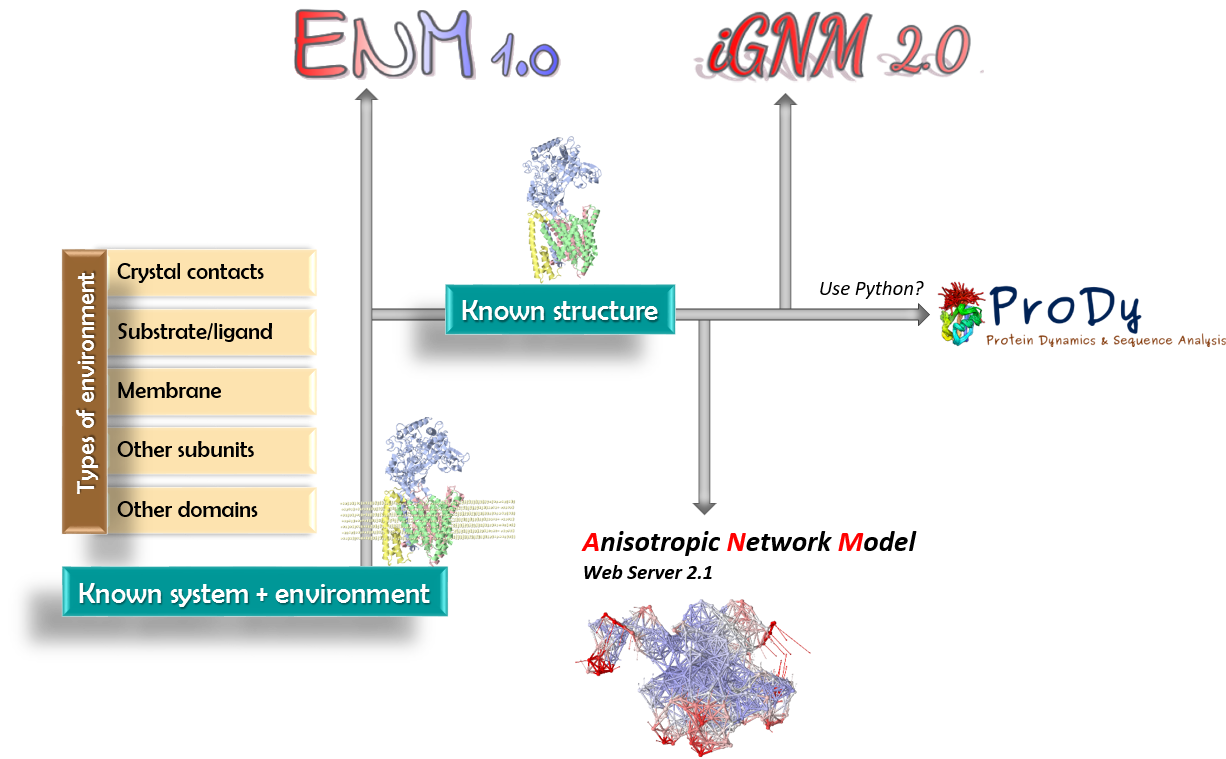 |
| See here a detailed flow chart describing how ENM 1.0 operates, the available options, methods used and type of outputs is provided. |

|
Reference: Li,H., Chang,YY, Lee,JY, Bahar,I. and Yang,LW. (2017) DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res., online published. Contact: The server is maintained by Dr. Hongchun Li in the Bahar Lab at the Department of Computational & Systems Biology at the University of Pittsburgh, School of Medicine, and sponsored by the NIH awards P30 DA035778 and #5P41GM103712-03 and the funding #104-2113-M-007-019 from MOST to the Yang lab at the National Tsing Hua University, Taiwan. For questions and comments please contact Hongchun Li.
|